The phage display
Introduction.
Phage display is a laboratory technique used for the high-throughput screening of protein- and peptide-binders. In this technique, a gene encoding a protein of interest (POI) is expressed in frame (i.e. as a fusion) with the phage coat protein gene, causing the phage to "display" the protein on its outside. The phage itself serves as a 'vessel' connecting the displayed phenotype and genotype. The displaying phages can then be screened against various bait molecules (proteins, peptides or DNA sequences) for specific interaction with them. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection. The most common bacteriophages used in phage display are M13 and FD filamentous phage, though T4, T7, and λ phage are also used.
General principle.
In the M13 phage display, the DNA encoding POI is expressed N-terminally with either pIII or pVIII gene, which encodes the minor and major coat proteins. The pIII gene is responsible for binding to E.coli via associating with the F' pilus thereby triggering infection. Certain E.coli strains are suitable for this purpose: TG1, SS320, ER2738, or XL1-Blue. Fusing POI to the pIII N-terminus ensures correct exposure of a target peptide for the interaction with a bait molecule. The POIs of sizes less than 50 amino acids largely don't affect the main function of the coat protein - phage infectivity. For larger POIs a "phagemid" vector is used (a simplified display construct vector) that encodes a fused version of the coat-protein with the intact coat protein (and other phage lifecyle-components) provided by a helper phage. Upon infection, intact pIII competes with POI-pIII fusion resulting in a large portion of the phage population not displaying POI and a small portion that would contain on average a single copy of POI-pIII per phage particle. Use of phagemid is beneficial in two ways: (1) smaller vector enables higher transformation efficiency hence higher genetic diversity libraries; (2) use of monovalent display of POI on phages often results in selection of higher affinity binders due to the absence of an 'avidity' effect.
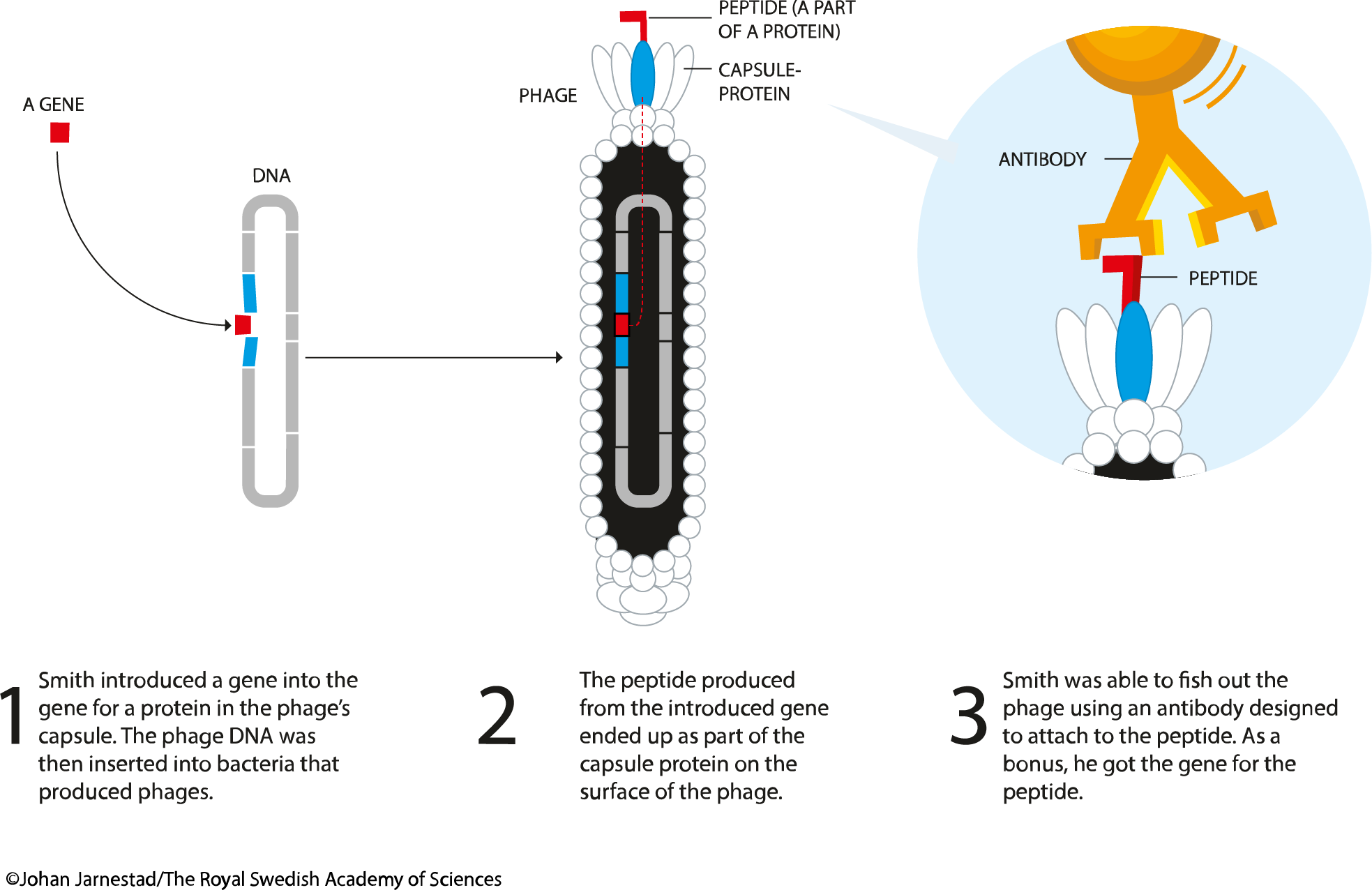
The bait molecules are typically immobilised on the surface of a microtiter plate. Phages that tightly interact with the bait will remain attached to the plate while the majority of the phage population will be washed away. The remaining phage are then eluted and amplified again in the next round of bacterial infection resulting in a phage mixture enriched with phages that bind the target molecule. This process is referred to as panning is typically repeated a number of times to establish high level of enrichment of phages encoding tight-binder POIs. Phage eluted in the final step can then be used to infect a suitable bacterial host, from which the phagemids are collected and subsequently analysed by NGS or Sanger sequencing coupled with ELISA.

The use of a helper phage can be eliminated by using 'bacterial packaging cell line' technology1. Elution is usually carried out by using trypsin to digest the bait protein and presented binding protein, thus releasing the bound phage, or by using low pH buffer to dissociate electrostatic interactions. Competitive elution can also be used if a known binder is available and binder for a specific region (e.g. a catalytic pocket) is desired.
General protocol.
A common protocol for the phage display would consist of the following steps: 1. Bait protein or DNA molecules are immobilized to the wells of a microtiter plate either chemically or via biotin:streptavidin interaction. 2. GOI sequences are expressed in the bacteriophage library as a fusion with a bacteriophage coat protein. 3. This phage-display library is added to the dish and after allowing the phage time to bind, the dish is washed. 4. Phage-displaying proteins that interact with the target molecules remain attached to the dish, while all others are washed away. 5. Attached phage may be eluted and used to create more phage by infection of suitable bacterial hosts. The new phage constitutes an enriched mixture, containing considerably less irrelevant phage (i.e. non-binding) than were present in the initial mixture. 6. Steps 3 to 5 are optionally repeated one or more times, further enriching the phage library for high-affinity binding phages. 7. Following further bacterial-based amplification, the DNA within the interacting phage is sequenced to identify the interacting proteins or protein fragments.
Challenges and requirements
Benefits.
The protocols are very well described and the techniques have been used for a long time now. The availability of different commercial kits makes this technique easy to be performed by non-experts. Phage display is quick, with four rounds of selection and screening feasible within two weeks.
Problems.
Given the biology of the phages, fast-growing phages will be enriched and peptides that are not relevant for the binding isolated. Also non-specific binders are often enriched because of the affinity of certain peptide/protein variants to the plastic of the titer plates or other supports used during the panning.
Level of complexity.
This technique is relatively easy, especially if starting from commercial kits. However, it is more complex if beginning from a different starting point and if rounds of diversification are needed. Materials and resources are the ones used in standard molecular biology techniques and instruments. However, to harvest phages, ultracentrifugation may be required. A plate reader is useful for reading ELISA plates, and the materials for ELISA needed for characterizing the binding of selected peptides can be expensive.
References and useful protocols
The best protocol to start with is the one present in the manual of the commercially available NEB kit for phage display:
These kits starts from random short peptide sequences, if the starting point is a non-random sequence then the target sequence should be cloned into M13 KE cloning phage following protocols also from NEB
Most of the relevant protocols for immobilization of the targets on the different solid support are also available on the NEB webpage
-
Chasteen L, Ayriss J, Pavlik P, Bradbury AR. Eliminating helper phage from phage display. Nucleic Acids Res. 2006;34(21):e145.https://doi.org/10.1093/nar/gkl772 ↩